Closed
Andrea Masotti, Bambino Gesù Children Hospital, Rome, Italy
3-years Project
Celiac Disease
Area: Genetics
- Grant: FC 018/2013
- Title: Novel biomarkers for the diagnosis of celiac disease and to monitor the adherence to the gluten-free diet.
- Topic: Celiac Disease, Genetics
- Duration: Triennial Project
- Principal Investigator: Andrea Masotti, Bambino Gesù Children Hospital, Rome
Publications originating from the Project:
- Felli C, Baldassarre A, Uva P, Alisi A, Cangelosi D, Ancinelli M, Caruso M, Paolini A, Montano A, Silano M, Vincentini O, Catassi C, Lionetti E, Gatti S, Ferretti F, Masotti A. Circulating microRNAs as novel non-invasive biomarkers of paediatric celiac disease and adherence to gluten-free diet. EBioMedicine. 2022 Feb;76:103851. doi: 10.1016/j.ebiom.2022.103851. Epub 2022 Feb 9. PMID: 35151110; PMCID: PMC8842006. https://pubmed.ncbi.nlm.nih.gov/35151110/
- Baldassarre A, Felli C, Prantera G, Masotti A. Circulating microRNAs and Bioinformatics Tools to Discover Novel Diagnostic Biomarkers of Pediatric Diseases. Genes (Basel). 2017 Sep 19;8(9):234. doi: 10.3390/genes8090234. PMID: 28925938; PMCID: PMC5615367. https://pubmed.ncbi.nlm.nih.gov/28925938/
- Felli C, Baldassarre A, Masotti A. Intestinal and Circulating MicroRNAs in Coeliac Disease. Int J Mol Sci. 2017 Sep 6;18(9):1907. doi: 10.3390/ijms18091907. PMID: 28878141; PMCID: PMC5618556. https://pubmed.ncbi.nlm.nih.gov/28878141/
The Study
PROJECT RATIONALE AND AIMS
Celiac disease (CD) is a systemic immune-mediated enteropathy triggered by dietary gluten. The only treatment is a strictly gluten-free diet (GFD) that leads to a symptomatic, serologic, and histological remission in most patients, but if it fails, other complications such as lymphoma can occur. The current gold standard for the diagnosis is a small-bowel biopsy together with positive serology, but only a small proportion of cases of CD are clinically recognized. Moreover, there are no clear guidelines for assessing the outcome or the adherence to the GFD. Thus, the discovery of new biomarkers is necessary not only for the diagnosis of CD, but also for the evaluation of the response to diet.
In the last few years many evidences demonstrated that during diseases (i.e. various types of cancer, diabetes) miRNAs circulating in serum originate from tissues or as a consequence of the inflammatory response. Our hypothesis is that circulating miRNAs can be used in combinations with other serological tests as less invasive biomarkers, respect to biopsy, for the diagnosis of CD and the response to the GFD.
The aim of this study is to identify predictive miRNAs as novel diagnostic markers for the diagnosis of CD and the response to GFD. Besides, by determining the altered expression levels of miRNAs in serum of patients with active CD and on GFD, we will identify miRNAs which are indicative of the response to GFD and consequently of the remission of small intestine damage, in order to avoid invasive bioptic procedures. Moreover we will correlate the expression levels of deregulated miRNAs with serological tests and with the grade of intestinal damage finally determining the diagnostic utility of selected circulating miRNAs.
RESEARCH PLAN AND RESULTS OBTAINED
This study was an observational prospective cohort study with a control group. A total number of 120 subjects ranging from 3 to 15 years belonging to three groups: CD (n=40), CD on GFD for more than six months (n=40), and CTRL (n=40) that were consecutively recruited at the Bambino Gesù Children’s Hospital of Rome at the Hepatology, Gastroenterology and Nutrition Department from January 2014 to December 2018. The inclusion criteria were: age (3-15 years), positivity to TGA-IgA and EMA-IgA, and a duodenal biopsy classified as Marsh score ≥ 3 or based on international guidelines (for CD group), and CD patients already on GFD for more than six months (for GFD group). Controls were selected among a group of children with no history/diagnosis of inflammatory or other autoimmune conditions, negative family history of CD, and negative serological antibodies for CD. The exclusion criteria were: the use of oral corticosteroids or hormones, presence of other gastrointestinal diseases or autoimmune conditions, diabetes, major psychiatric disorders and physical impairment limiting physical activity.
EXPERIMENTAL DESIGN AND METHODOLOGIES
Blood was collected into serum separator tubes (BD Vacutainer ® SST™ II Advance). Samples were processed within 2h from blood withdrawal. Serum was obtained after centrifugation of the tubes at 1800 g for 15 min at room temperature. Total RNA was isolated by using the Plasma/Serum Circulating and Exosomal RNA Purification kit (Norgen) from 250 μl (for qPCR) or 500 μl (for small RNA sequencing) of serum depending on the particular analysis, according to manufacturer’s instructions. RNA was stored at −80°C until use. Circulating miRNAs were sequenced by using a small RNA sequencing protocol. Deregulated miRNAs detected by RNA sequencing were validated in the independent set by qPCR. The analyses were performed by using Serum/Plasma Focus miRNA PCR Panel I+II (Exiqon) which assessed 179 miRNAs with specific miRNA locked nucleic acid (LNA)™ PCR primers (Exiqon).
POTENTIAL PITFALLS AND CAVEATS
The main problem was the isolation of a good quality and high-yield RNA sample to be used in downstream analyses. We successfully adopted a protocol that we optimized in our laboratory.
CONCLUSIONS AND DISCUSSION
In this prospective observational study we investigated the presence of circulating miRNAs in the sera of patients affected by celiac disease or adhering to a gluten-free diet (GFD) by employing RNA-Seq and qPCR analyses that allowed us to identify potential biomarkers of disease and progression. We identified three miRNAs (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) that could be employed successfully not only to identify patients with biopsy-confirmed celiac disease, but also affected patients with low levels of TGA-IgA (referred as ‘borderline’ patients) (Fig.1-3). These three miRNAs are uniquely expressed in the sera of CD patients and may have an active role in regulating many important biological processes implicated in CD disease as their potential gene targets belong to apoptotic processes, cell cycle phase transitions and other immune-related processes. The use of a linear model containing information of the expression levels of circulating miRNAs within the various groups, can further improve the sensitivity of the method, and allowed to obtain a sensitivity of 68.8% and a specificity of 94.7 (AUC=0.867) for the diagnosis of CD patients (Fig.2). For the diagnosis of CD in borderline patients, the model was able to reach a sensitivity of 84.6% and a specificity of 84.2 (AUC=0.883). The model of the three miRNAs can identify patients under GFD with a sensitivity of 73.9% and a specificity of 96.9% (AUC=0.909), thus representing an innovative way to monitor patient’s adherence to diet (Fig.3). Nowadays, no other methods can allow to obtain this kind of information objectively, nor other analytical techniques or serological biomarkers are available yet to follow the adherence to a gluten-free diet for CD patients. Therefore, we strongly support the use of circulating miRNAs as a supplementary tool for the diagnosis of CD without recurring to intestinal biopsy, a procedure that, especially for children, may result quite invasive and not very tolerated.
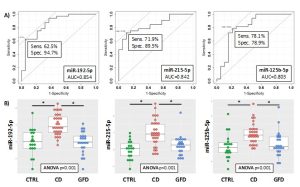
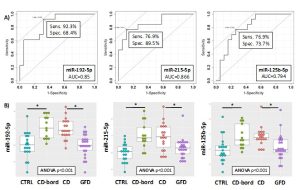
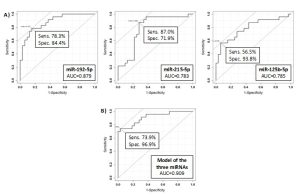
Figure 1. ROC curves for the discrimination of CD patients compared to CTRL (A) and DotPlots (B) of miR-192-5p, miR-215-5p and miR-125b-5p for the three groups of patients (CTRL, CD and GFD). The asterisk indicates a significant difference (p<0.001).

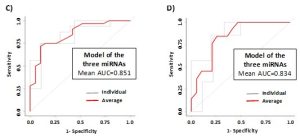
Figure 2. ROC curves for the discrimination of borderline CD patients (CD-bord) compared to CTRL (A) and DotPlots (B) of miR-192-5p, miR-215-5p and miR-125b-5p for the four groups of patients (CTRL, CD-bord, CD and GFD). The asterisk indicates a significant difference (p<0.001). ROC curves of the model obtained by using the three miRNAs (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) and the two-fold cross-validation for the diagnosis of CD patients compared to controls (C) and borderline CD patients compared to controls (D). Individual folds and average are colored in gray or red, respectively.

Figure 3. ROC curves for the discrimination of CD patients under a gluten-free diet (GFD patients) compared to CD patients (A) and ROC curve of the model obtained by using the three miRNAs (i.e., miR-192-5p, miR-215-5p and miR-125b-5p) for the discrimination of GFD patients compared to CD patients.
