closed
Mariavittoria Laezza, Institute of Genetics and Biophysics - CNR, Naples, Italy
3-years Project
Celiac Disease
Area: Genetics
Grant: 006/2019
- Title: Regulation of HLA DQ2.5 genes in patients with active celiac disease and patients on gluten free diet.
- Duration: Triennial Project
- Principal Investigator: Mariavittoria Laezza, Institute of Genetics and Biophysics – CNR, Naples, Italy
- Tutor (Head Lab): Giovanna Del Pozzo,Institute of Genetics and Biophysics – CNR, Naples, Italy
Publications originating from the Project
- Farina F, Pisapia L, Laezza M, Serena G, Rispo A, Ricciolino S, Gianfrani C, Fasano A, Del Pozzo G. Effect of Gliadin Stimulation on HLA-DQ2.5 Gene Expression in Macrophages from Adult Celiac Disease Patients. Biomedicines. 2021 Dec 29;10(1):63. doi: 10.3390/biomedicines10010063. PMID: 35052743; PMCID: PMC8773327. https://pubmed.ncbi.nlm.nih.gov/35052743/
- Del Pozzo G, Farina F, Picascia S, Laezza M, Vitale S, Gianfrani C. HLA class II genes in precision-based care of childhood diseases: what we can learn from celiac disease. Pediatr Res. 2021 Jan;89(2):307-312. doi: 10.1038/s41390-020-01217-4. Epub 2020 Oct 29. PMID: 33122841. https://pubmed.ncbi.nlm.nih.gov/33122841/
- In preparation: Laezza M et al. – “Gliadin antigen presentation and inflammation during gluten free diet”.
THE STUDY
Project rationale and aims
The genetic susceptibility to develop Celiac Disease (CD) is strongly associated with HLA class II DQA1*05 and DQB1*02 genes encoding for DQ2.5 molecules, carried by 95% of patients. The DQ2.5 heterodimers, expressed on Antigen Presenting Cells (APCs), present gluten peptides to CD4+ T cells that orchestrate the inflammatory cascade in CD patients. Although it has been demonstrated that the risk to have CD is dependent on the number of DQA1*05 and DQB1*02 alleles, recent data showed that the magnitude of the anti-gluten T cell immunity is shaped by their expression on APCs. Previous publications demonstrated that the strength of pathogenic CD4+ T cell response to dietary gluten is primarily determined by the expression of HLA class II CD risk alleles in professional APCs, and not by the number of risk genes carried.
The general aim of my work was the assessment of HLA-DQ2 expression in APCs from patients with active disease, patients in remission at gluten-free diet (GFD) and healthy subjects. We focused our analysis on three group of patients carrying heterozygous haplotypes: DR3/DR7, DR3/DR5 and DR5/DR7, associated to a different risk of pathology.
Research plan and results obtained
We enrolled volunteer patients at the Department of Gastroenterology of the Federico II University of Naples. The subjects enrolled in the study were affected by active celiac disease at diagnosis time and celiac patients on gluten-free diet with negative serology.
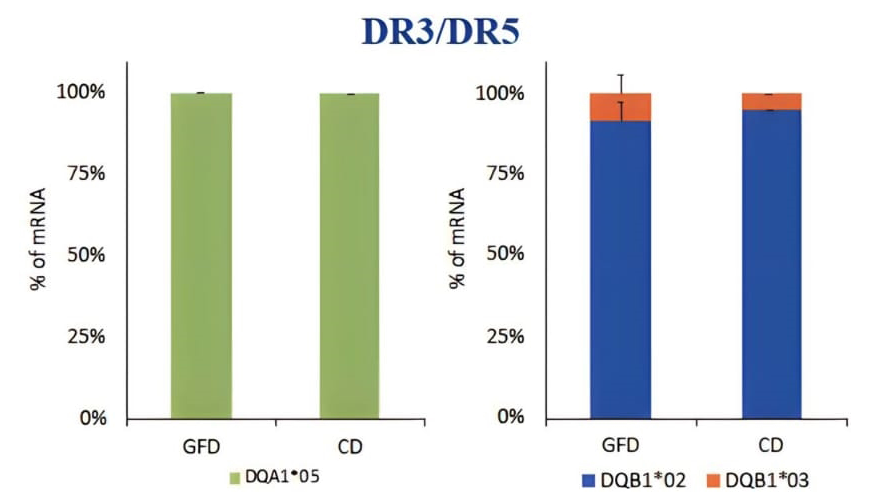
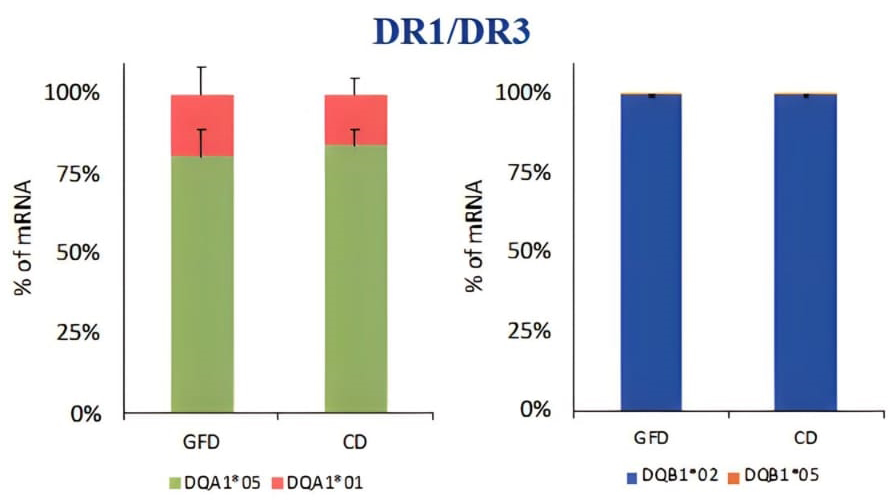
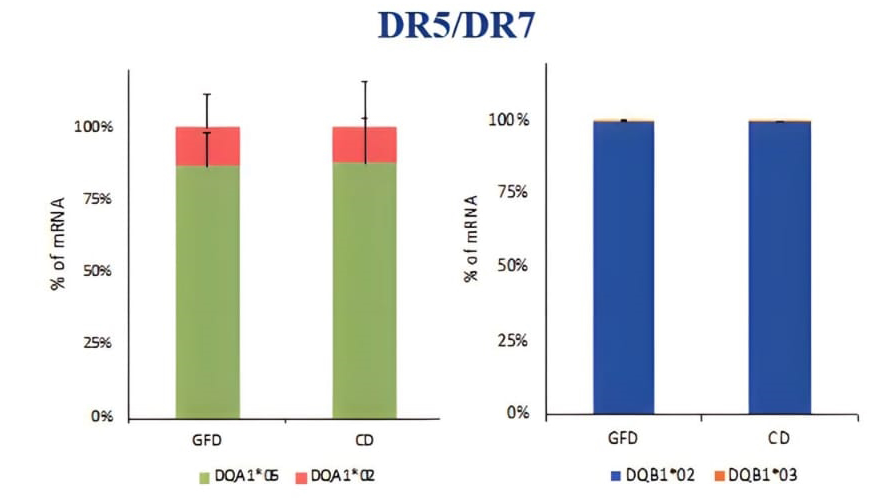
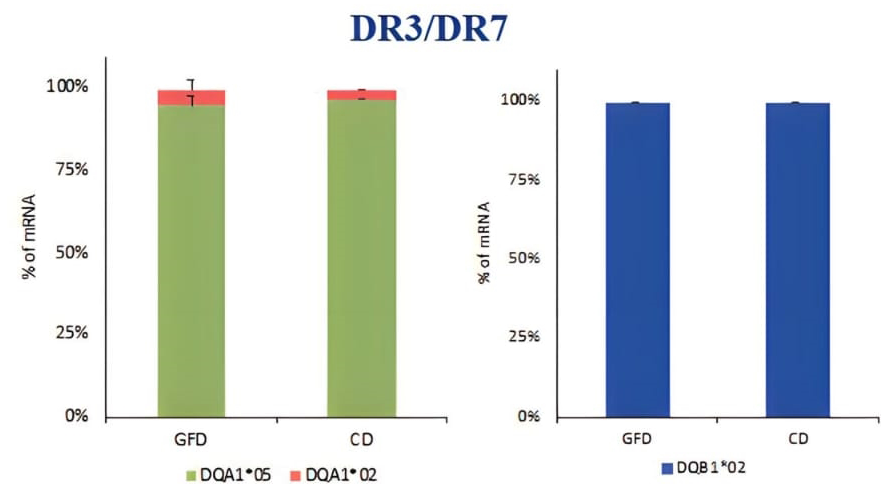
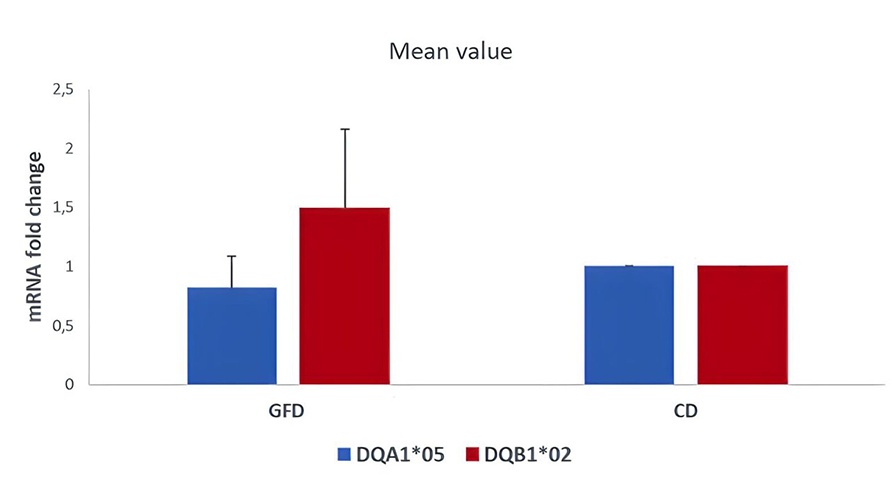
PBMCs (peripheral blood mononuclear cells) was a heterogeneous cell population containing many APC such us monocytes, dendritic cells, macrophages, and B cells expressing HLA class II molecules involved in the antigen presentation. PBMC were prepared for DNA and mRNA extraction. The HLA genotyping is essential for the selection of primers specific for each allele to be used for qPCR. After we identified the alleles carried by patients, we measured the HLA-DQ2 mRNA expression by absolute qPCR and we found that the amount of DQA1*05 and DQB1*02 mRNAs, transcribed by risk alleles, is significantly higher than the mRNA transcribed by HLA-DQ alleles not associated with celiac disease, in all samples analysed (Figure 1). Then, we compared the expression of DQA1*05 and DQB1*02 alleles between patients with active celiac disease and patients on GFD, regardless of genotypes, and we observed that there are not significant differences in the expression of the risk alleles (Figure 2). This result demonstrated that the HLA-DQ2 genes are expressed at same level and APC from both group of patients have the same capability to present gliadin antigenic peptides.
Figure 1. Expression of the DQA1*05 and DQB1*02 risk alleles compared to alleles not associated with the disease, expressed as a percentage of mRNA.
———————————————————————————–
Figure 2. Expression of DQ risk alleles DQAI*05 and DOB I *02 in patients with acute CD and on GFD, regardless of genotype.
Experimental design and methodologies
We enrolled volunteer patients at the Department of Gastroenterology of the Federico II University of Naples, during the first access in the clinic at diagnosis time or during the follow up after GFD. Our co-worker clinicians get 10 mL of blood and PBMCs were isolated from samples by density gradient centrifugation over Ficoll-Paque. After centrifugation, the mononuclear cells (lymphocytes and monocytes) were found in a whitish ring at interface between Ficoll and plasma, and after recovering, these cells were counted and washed, and eventually stored at -80°C for later experiments. To perform the RNA quantization by qPCR we need to know the HLA genotype of patients. The HLA genotype of patients was performed with the BioDiagene kit, a commercial diagnostic kits for the detection of HLA class II alleles associated with CD and for the direct identification of samples homozygous for DQB1*02 allele. The test is based on a preliminary lysis of a small amount of blood, DNA amplification and agarose gel electrophoresis.
Total RNA was extracted by PBMCs by Trizol reagent and used for reverse transcriptase reactions with Superscript III. cDNA was quantified by qRT-PCR using SYBR Green and the BIORAD Real-Time PCR Detection System. An absolute mRNA quantification was carried out with allele-specific pairs of primers, through the synthesis of a cDNA standard.
Potential pitfalls and caveats
The main problem of work was the reduced number of healthy volunteers carrying the HLA-DQ2 genotype that we enrolled in the study. As consequence we limited our analysis to the DQ2.5 gene expression among patients with acute disease and patients at GFD.
Conclusions and discussion
The first achievement of the project was to confirm the elevated expression of HLA-DQ2 risk alleles (DQA1*05, DQB1*02), compared to alleles not associated with celiac disease, in the circulating APC with heterozygous HLA.DQ2 genotype. Moreover, we demonstrated comparable expression of DQ2 in APC from patients with active disease and patients on GFD, independently from their genotype. This result demonstrated that the autoimmune response by gliadin specific activated CD4 T lymphocytes is determined by the presence of the antigen and not caused by the density of DQ2 molecules expressed by APC.
The high expression of risk alleles is a finding also common to other autoimmune pathologies as summarized in the review Pediatric Research (Farina, F. et al., 2019; Pisapia, L. et al., 2019; Cavalli, G. et al., 2016; Hayashi, M. et al., 2016; Raj, P. et al.; 2016). This literature and the previous publications by Del Pozzo lab strongly indicated that the high expression of HLA-DQ2 alleles contribute to the risk of developing coeliac disease. Instead, the present work demonstrated that the HLA-DQ2 expression is comparable in PBMC from patients with active disease and patients on GFD. The disease recovery does not affect HLA-DQ2 expression and the absence of CD4 +T cell activation is a consequence of lack of antigen.
