closed
Michele Sallese, University of Chieti-Pescara, Italy
3-years Project
Gluten Sensitivity
Area: Genetics
- Grant: FC 046/2013
- Title: Identification of gene expression profiling to the diagnosis of celiac disease and non-celiac gluten sensitivity
- Topic: Gluten Sensitivity, Genetics
- Duration: Triennial Project
- Principal Investigator: Michele Sallese, University ‘G. D’annunzio’ Chieti-Pescara
Publications originating from the Project:
- Efthymakis, K.; Clemente, E.; Marchioni, M.; Di Nicola, M.; Neri, M.; Sallese, M., An Exploratory Gene Expression Study of the Intestinal Mucosa of Patients with Non-Celiac Wheat Sensitivity. Int J Mol Sci 2020, 21 (6). https://pubmed.ncbi.nlm.nih.gov/32183058/
- Clemente, E.; Efthymakis, K.; Carletti, E.; Capone, V.; Sperduti, S.; Bologna, G.; Marchisio, M.; Di Nicola, M.; Neri, M.; Sallese, M., An explorative study identifies miRNA signatures for the diagnosis of non-celiac wheat sensitivity. PLoS One 2019, 14 (12), e0226478. https://pubmed.ncbi.nlm.nih.gov/31834915/
The Study
PROJECT RATIONALE AND AIMS
Non-celiac gluten/wheat sensitivity (NCGS/NCWS) is a syndrome dependent on gluten/wheat ingestion that presents both intestinal and extra-intestinal symptoms.
NCWS clinical signs include alternate bowel habits, bloating, abdominal pain, fatigue, foggy mind, limbs numbness, dermatitis, joint and muscle pain. These symptoms largely overlap with those of other wheat-related disorders, including celiac disease and wheat allergy. Given the lack of NCWS specific biomarkers, the most advanced diagnostic algorithm implies the exclusion of confounding pathologies and the assessment of symptoms during a gluten challenge.
To date we still know little about NCWS inducing factors, possible genetic predisposition and pathological mechanisms, besides the fact that they probably differ from celiac disease and wheat allergy. Previous studies aimed at identifying the features of NCWS reported the downregulation of Foxp3, the upregulation of toll-like receptor 2 and claudin-4, a modest production of IFNγ and an increase of intraepithelial lymphocytes in the intestinal mucosa. In addition, lipopolysaccharide (LPS)-binding protein and fatty acid-binding protein 2 were shown to be increased in the plasma of these patients.
Overall, NCWS presents minor signs of intestinal pathology, at least by using the classical approaches. We hypothesized that subtle changes of multiple miRNAs or genes might be sufficient to cause the intestinal discomfort, therefore we evaluated differences in miRNA and gene expression in NCWS patients and non-NCWS patients. This analysis aimed to shed light on the molecular mechanism that regulates this disorder and to identify possible biomarkers of NCWS that could be helpful to the diagnosis.
RESEARCH PLAN AND RESULTS OBTAINED
To date, the diagnosis of NCWS relies on symptoms and exclusion of confounding diseases, since biomarkers are not yet available. Here, the expression levels of selected miRNAs were examined in duodenal biopsies and peripheral blood leukocytes collected from newly diagnosed patients with NCWS and, as controls, from patients with celiac disease and gluten-independent gastrointestinal problems. This screening lead to the identification of seven miRNA (hsa-miR-19b-3p, hsa-miR-19a-3p, hsa-miR-186-5p, hsa-miR-17-5p, hsa-miR-145-5p, hsa-miR-30e-5p, hsa-miR-143-3p) which are significantly overexpressed in the intestinal mucosa of NCWS patients in comparison to control patients. Notably, six of these miRNA were found to be overexpressed also in peripheral blood leukocytes (PBL) of NCWS patients. Principal components analysis (PCA) of the miRNA expressions showed that the first component (PC1) summarises most of the variance and correlates with the NCWS status. The PC1 performance, as a classifier, estimated by Receiver Operating Characteristic (ROC) curves, indicated 74-76% of probability to classify these patients (Figure 1).
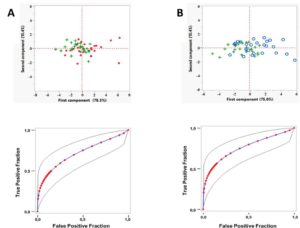
Figure 1. A principal component analysis of miRNA expression levels can classify patients with NCWS.
(Upper panels) A principal component analysis was performed on the ΔCt values for each differentially expressed miRNA. Plot of the first and second component is shown. Green + represent the NCWS group, red dots represent the control group (A) and blue circles represent the celiac group (B). (Lower panels) Fitted ROC curve (blue line) of the first principal component data (red dots). Plotting of the true positive rate versus false positive rate as defined by the PC1 values. AUC = 0.74 ± 0.0702 (p<0.001, Wilcoxon U test) for the comparison NCWS vs controls (A). AUC = 0.76 ± 0.0680 (p<0.001, Wilcoxon U test) for the comparison NCWS vs celiac (B). Grey lines: 95% confidence interval of the fitted ROC curve.
Next, we carried out a genome wide expression analysis on RNA extracted from intestinal mucosa of NCWS patients, and age-matched controls. We revealed the presence of 300 differentially expressed (DE) transcripts, the majority of which were non-coding RNA. PCA of these DE genes indicated that the first component is sufficient to summarise the characteristics of NCWS and have the appropriate power to segregate NCWS from controls. Furthermore, we reported a classification of the transcripts that contribute the most to the NCWS status (Figure 2). Our findings suggest that expression profiling may be helpful in positive diagnosis and/or exclusion of NCWS in symptomatic subjects. Finally, protein-coding transcripts analyses suggested that innate immune response plays a role in NCWS.
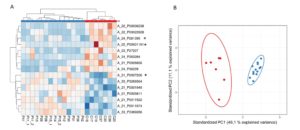
Figure 2. Heat map and PCA representation of the most predictive transcripts.
(A) Reddish and bluish colours represent upregulated and downregulated transcripts respectively. The columns of the heat map represent the patients organised according to Canberra distance algorithm while the rows represent the transcripts organised according to correlation distance. The code shown below the columns identifies a specific patient. (B) Scatter plot of the first two components (PC1 and PC2) summarising the expression levels of the transcripts selected by LASSO. Red and blue dots represent controls and NCWS patients respectively.
EXPERIMENTAL DESIGN AND METHODOLOGIES
Patients expressing the HLA-DQ2/DQ8 haplotypes, showing positive celiac disease serology (IgA anti-tTG and/or anti-EmA), and villous atrophy at histology were enrolled as celiac. Patients presenting with gastrointestinal symptoms, negative celiac disease serology and preserved mucosal architecture, were prescribed a gluten-free diet for a 6 week period, after which persistently symptom-free subjects were reintroduced to dietary wheat protein. The recurrence of symptoms prompted the final diagnosis of NCWS. Finally, patients with gluten-independent dyspeptic symptoms who required endoscopic examination were also recruited as controls. Total RNA was extracted from duodenal biopsies and PBL. The expression of selected miRNAs was evaluated by quantitative real time PCR, while genome wide gene expression was evaluated by microarray analysis. Differentially expressed miRNA and genes were identified by statistical analysis.
POTENTIAL PITFALLS AND CAVEATS
The proposed research, although based on well-established approaches, could present potential pitfalls. An insufficient number of patients could enter the study within the scheduled time. To overcome this problem, we have extended the recruitment period to enrol the necessary number of patients. An extremely limited number of differentially expressed genes could be identified by the qPCR analysis of selected genes (original experimental plan); and this was the case. Therefore, we extended the number of genes investigated by qPCR, but despite these measures, we were unable to identify differentially expressed genes. Finally, we decided to carry-on a genome wide expression analysis using a microarray approach.
CONCLUSIONS AND DISCUSSION
The clinical manifestations and the pathogenic mechanisms of the recently defined NCWS are still little understood and the diagnosis is not supported by laboratory tests. We investigated miRNA and gene expression profiles of NCWS with the aim of shedding light on the molecular mechanisms governing this pathology and eventually identifying effective biomarkers helpful to the diagnosis. In the intestinal mucosa of patients affected by NCWS we identified a few miRNAs whose expression is higher in comparison to control patients affected by gluten-independent dyspeptic symptoms and celiac disease. Furthermore, the intestinal mucosa of NCWS patients present 300 differentially expressed transcripts in comparison to dyspeptic controls. This study provided the first evidence that NCWS patients can be genetically defined. The relevance of our finding is underscored by the current lack of biomarkers and laboratory tests useful to validate the diagnosis of NCWS. Differentially expressed genes and/or miRNA patterns combined with serological and histological exclusion of celiac disease, might represent a future marker for positive diagnosis of this syndrome. However, validation on larger groups of patients is needed to introduce this type of biomarker into the clinic.
An analysis of the functional role of the dysregulated genes suggested that NCWS may result from a pathological immune response, especially of the innate branch. Furthermore, non-coding RNA could play an important role in the pathogenesis of NCWS.
